An electronic chip that can analyze blood and other clinical samples for infectious bacteria with record-breaking speed
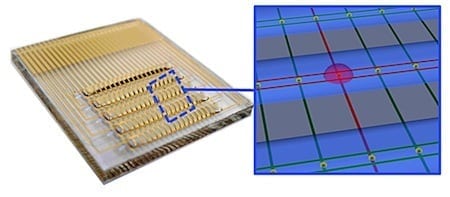
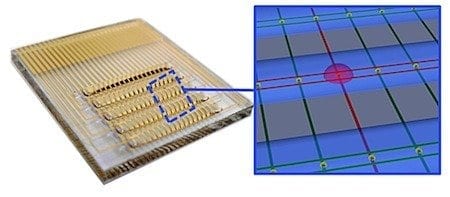
A U of T team – including researchers from Electrical and Computer Engineering and the Institute of Biomaterials & Biomedical Engineering – has created an electronic chip that can analyze blood and other clinical samples for infectious bacteria with record-breaking speed.
Life-threatening bacterial infections cause tens of thousands of deaths every year in North America but current methods of culturing bacteria in the lab can take days to report the specific source of the infection, and even longer to pinpoint the right antibiotic that will clear the infection.
The new technology, reported in the journal Nature Communications , can identify the pathogen in a matter of minutes, and looks for many different bacteria and drug resistance markers in parallel, allowing rapid and specific identification of infectious agents.
“Overuse of antibiotics is driving the continued emergence of drug-resistant bacteria,” said Shana Kelley (Pharmacy and Biochemistry), a senior author of the study. “A chief reason for use of ineffective or inappropriate antibiotics is the lack of a technology that rapidly offers physicians detailed information about the specific cause of the infection.”
The researchers developed an integrated circuit that could detect bacteria at concentrations found in patients presenting with a urinary tract infection. “The chip reported accurately on the type of bacteria in a sample, along with whether the pathogen possessed drug resistance,” explained Chemistry PhD student Brian Lam, the first author of the study.
A U of T team – including researchers from Electrical and Computer Engineering and the Institute of Biomaterials & Biomedical Engineering – has created an electronic chip that can analyze blood and other clinical samples for infectious bacteria with record-breaking speed.
Life-threatening bacterial infections cause tens of thousands of deaths every year in North America but current methods of culturing bacteria in the lab can take days to report the specific source of the infection, and even longer to pinpoint the right antibiotic that will clear the infection.
The new technology, reported in the journal Nature Communications , can identify the pathogen in a matter of minutes, and looks for many different bacteria and drug resistance markers in parallel, allowing rapid and specific identification of infectious agents.
“Overuse of antibiotics is driving the continued emergence of drug-resistant bacteria,” said Shana Kelley (Pharmacy and Biochemistry), a senior author of the study. “A chief reason for use of ineffective or inappropriate antibiotics is the lack of a technology that rapidly offers physicians detailed information about the specific cause of the infection.”
The researchers developed an integrated circuit that could detect bacteria at concentrations found in patients presenting with a urinary tract infection. “The chip reported accurately on the type of bacteria in a sample, along with whether the pathogen possessed drug resistance,” explained Chemistry PhD student Brian Lam, the first author of the study.
The Latest Bing News on:
Pathogen Identification
- Loss of large herbivores affects interactions between plants and their natural enemies, study showson May 7, 2024 at 7:33 am
Insects and microorganisms that feed on plants, cut up leaves, modify leaf tissue or produce leaf spots and other kinds of damage, are usually known as pests and considered harmful, yet interactions ...
- Key role found for gut epithelial cells in the defense against deadly diarrheal infectionson May 7, 2024 at 7:14 am
Intestinal epithelial cells line the inner wall of the gut, creating a barrier against dangerous bacteria like enteropathogenic E. coli that seek to attach to and destroy this barrier. Such pathogens ...
- Faces of Food Safety: Meet Dayna M. Harhay of the Animal Research Centeron May 5, 2024 at 5:00 pm
identification, and prevention of Salmonella bacteria and other foodborne pathogens that can be associated with beef products. Isn’t Salmonella a Chicken Problem? Harhay smiles as she looks back at ...
- Spanish Startup DeepUll Unveils MDx Platform, Direct-From-Blood Sepsis Assay at ESCMID Globalon April 29, 2024 at 3:00 am
DeepUll showcased its UllCore platform and UllCore BSI Test, a one-hour multiplex PCR assay targeting 95 percent of sepsis-causing pathogens and multiple antibiotic resistance genes.
- Study rethinks immune system strategies to boost vaccine effectivenesson April 28, 2024 at 9:52 pm
A recent Cell journal review explores key immunological assumptions to enhance our understanding of vaccine design, autoimmune responses, and immune system pathology, aiming to refine intervention ...
- The JRC explains: technologies for fighting airborne germs - PCR, sequencing, and the reston April 26, 2024 at 3:10 am
During the COVID-19 pandemic, most of us made a rather intimate acquaintance with some of these technologies. And many of us are still confused about ...
- How immune cells communicate to fight viruses: New mouse model enables identification of chemokine producers and sensorson April 25, 2024 at 8:20 am
Chemokines are signaling proteins that orchestrate the interaction of immune cells against pathogens and tumors. To understand this complex network, various techniques have been developed to identify ...
- deepull unveils 1 hour, direct-from-blood multiplex PCR test for 95% of sepsis-causing pathogens at ESCMID 2024on April 25, 2024 at 12:28 am
UllCORE is designed to transform life-saving clinical decision making for sepsis patients Barcelona, Spain – 25 April 2024 – deepull, a medical diagnostics company developing culture-free diagnostic ...
- Federal legislation unveiled to address Reedley lab issues: ‘You don’t want deadly pathogens next door’on April 23, 2024 at 3:37 pm
Congressman Jim Costa unveiled legislation on Tuesday to address gaps in federal law in the regulation of highly infectious agents and high-containment laboratories, spurned on by the discovery of an ...
- deepull appoints Kimberle Chapin, MD, as Chief Medical Officer and Wade Stevenson as Chief Marketing and Sales Officeron April 17, 2024 at 11:30 pm
Kimberle Chapin, MD, as Chief Medical Officer and Wade Stevenson as Chief Marketing and Sales OfficerKimberle Chapin brings ...
The Latest Google Headlines on:
Pathogen Identification
[google_news title=”” keyword=”Pathogen Identification” num_posts=”10″ blurb_length=”0″ show_thumb=”left”]
The Latest Bing News on:
Bacterial infection detection
- Avian metapneumoviruses are difficult to controlon May 7, 2024 at 5:00 pm
Edited version of a presentation by Silke Rautenschlein, DVM, PhD, University of Veterinary Medicine Hannover, Germany, during the 2024 Western Poultry Disease Conference] While the clinical signs of ...
- Why Your Breast Hurts in a Specific Spoton May 7, 2024 at 2:28 pm
Experiencing pain or discomfort in the breasts is a common concern for many individuals. Several factors, ranging from hormonal fluctuations to more serious conditions like breast cancer, can ...
- Mastiff's ear care: How to prevent infections in your dogon May 7, 2024 at 12:57 am
Each week, clean your Mastiff's ears with a veterinarian-recommended solution. Use a cotton ball or a soft cloth to gently wipe the outer ear, steering clear of the ear canal to avoid damage. This ...
- Are these dog infections the most deadly?on May 5, 2024 at 5:49 am
Seek prompt veterinary care if your dog shows signs of respiratory infection to prevent further complications.
- The Cautionary Parable of the Bacteria that Cause Lyme Diseaseon May 4, 2024 at 4:03 am
John Van Sloten says creatures such as ticks that spread Lyme disease can serve as a parable for spiritual parasites -- they are outside intruders that, if we let them, can rob us of life. There’s a ...
- Urine Chemokines May Improve Noninvasive Detection of Kidney Transplant Rejectionon May 3, 2024 at 5:42 am
Incorporating the chemokines in a model strongly detected clinically relevant rejections warranting biopsy and, if confirmed, antirejection treatment.
- Cats suffer H5N1 brain infections, blindness, death after drinking raw milkon April 29, 2024 at 3:41 pm
In a study published today in the journal Emerging Infectious Diseases, researchers in Iowa, Texas, and Kansas found that the cats had H5N1 not just in their lungs but also in their brains, hearts, ...
- Pneumonia: What to Knowon April 27, 2024 at 5:00 pm
Pneumonia is a lung infection that can range from ... Atypical pneumonia is caused by bacteria that are hard for doctors to detect with standard methods, such as mycoplasma and legionella.
- WSU researchers expose ‘bacterial vampirism’ in some foodborne pathogenson April 26, 2024 at 5:00 pm
These bacteria are known culprits behind bloodstream infections, particularly among individuals with ... pinpointing serine as a key chemical cue from blood that bacteria detect and consume. Siena ...
- Woman died of aggressive lung infection - inqueston April 26, 2024 at 9:33 am
A mum-of-three died due to an "aggressive" bacterial infection which caused lung damage she could not recover from, a coroner has concluded. Hannah Plumb, 32, died at University Hospital Coventry in ...
The Latest Google Headlines on:
Bacterial infection detection
[google_news title=”” keyword=”bacterial infection detection” num_posts=”10″ blurb_length=”0″ show_thumb=”left”]