Autonomously growing synthetic DNA strands
Synthetic biologists and nanobiologists are re-purposing DNA, the hereditary material present in nearly all the body’s cells, as a smart and stable self-assembling material to build nanofactories, drug-delivering nanostructures and molecular devices that can sense their environment and respond in different ways by, for example, detecting inflammation in the body or toxins in the environment. These nanoscale applications often involve the synthesis of large sequences comprising thousands of the building blocks that DNA is made of, known as the A, T, C and G nucleotide bases, which can be further folded and structured due to the specific base-pairing abilities between As and Ts, and Cs and Gs, respectively.
However, so far, researchers do not have tools at their disposal that would allow larger single-stranded sequences to autonomously grow and then join each other end-to-end following a molecular design plan, a capability that could generate structures and devices with diverse capabilities.
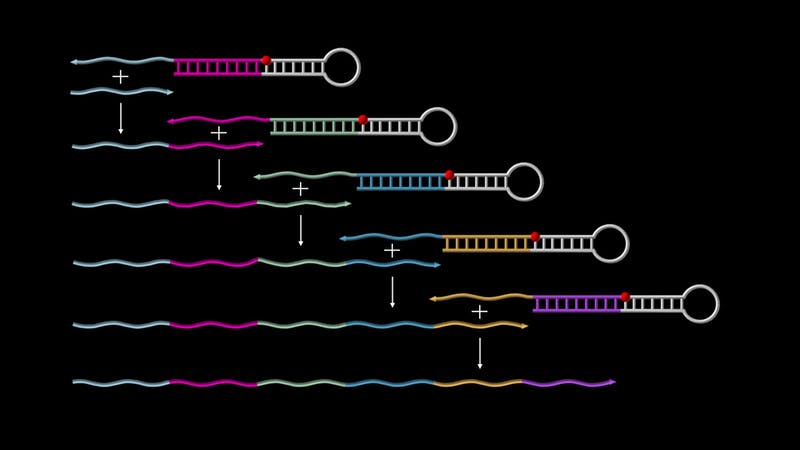
Published today in Nature Chemistry, research by Peng Yin at Harvard’s Wyss Institute for Biologically Inspired Engineering provides a broadly applicable solution to this problem. Yin and his team have developed a method that allows pre-designed sequences of DNA to autonomously grow and concatenate along specific assembly routes, hence providing the basis for a new generation of programmable molecular devices. Putting their new concept of so-called ‘Primer Exchange Reaction’ (PER) cascades to the test, they successfully engineered a first set of devices with diverse functions, such as self-building DNA-origami and DNA nanostructures that sense, amplify, record or logically evaluate environmental signals.
Past methods produced identical copies of a fixed smaller sequence, but they are unable to append different synthesized sequences to each other in defined patterns to generate larger assemblies autonomously without user-mediated intervention. “The autonomous and programmable features that PER cascades offer could engender an entirely new generation of programmable molecular devices and applications and close gaps in design efforts, for which many moving parts already exist,” said Wyss Institute Core Faculty member Peng Yin, Ph.D., who led the study and is also Professor of Systems Biology at Harvard Medical School (HMS). “We provide proof-of-concept data for PER in a diverse range of state-of-the-art synthetic biology applications that clearly highlight the technology’s broad potential.”
The Wyss Institute’s team used the new concept to design a series of such PER DNA transcripts for very diverse applications, including the autonomous synthesis of large DNA nanostructures known as DNA-origamis, and synthetic biology approaches, in which the synthesis of a DNA transcript hinges on a trigger, such as a cancer-associated small micro RNA. Their PER approach can even generate DNA transcripts resulting from a logically evaluated combination of different triggers, similar to RNA Ribocomputing Devices that Yin’s team published earlier this year.
Interestingly, PER DNA transcripts can become catalytic themselves, being able to cut an arbitrary target RNA, become fluorescently labeled probes that amplify the presence of a particular molecular stimulus, or “molecular recorders” that faithfully indicate the order in which certain molecular signals appear in their environments.
To start the PER cascade, two basic components are needed. One is called a “catalytic DNA hairpin mediator”, which is a single-stranded DNA molecule that partially pairs up with itself to form a hairpin structure with a short overhanging single strand. This overhang is designed to capture the PER cascades’ second component, the “primer,” which contains a region that is complementary to the overhang. Through a series of elongation and displacement reactions, the primer is extended with a sequence provided by the catalytic hairpin mediator and then expelled. This frees up the catalytic hairpin mediator to cascade the next round of the process, either by capturing a new starting primer or the already elongated primer — and so forth.
These complex synthesis pathways proceed autonomously, comparable to a molecular robot performing a given task, and at a single temperature, which makes the technology very robust. “The approach gives us tremendous creative freedom: we can not only synthesize the same piece of DNA again and again as new additions of a growing sequence, but we can also vary the types of DNA sequences to be appended simply by changing the composition of catalytic hairpin DNAs and primers in the mix while the assembly is ongoing. This allows us to have the synthesis branch off into different directions and to intricately pattern the composition of the final DNA transcript,” said the study’s first author Jocelyn Kishi, who as a National Science Foundation (NSF) Graduate Research Fellow at HMS works on Yin’s Wyss Institute team. “We are now working toward implementing PER cascades for a variety of applications, including molecular recorders, sophisticated diagnostics, and tissue imaging. We also hope that someday these systems can be used in living cells as devices that can record events or re-program cell behavior in specific ways,” said Kishi.
“This new advance that shows how DNA molecules can be programmed to self-assemble into specifc 3D structures and carry out predefined functions and tasks represents a major step forward in the field of Molecular Robotics, and provides a glimpse into the future of devices for both medical and non-medical applications,” said Wyss Institute Founding Director Donald Ingber, M.D., Ph.D., who is also the Judah Folkman Professor of Vascular Biology at HMS and the Vascular Biology Program at Boston Children’s Hospital, as well as Professor of Bioengineering at the Harvard John A. Paulson School of Engineering and Applied Sciences (SEAS).
Learn more: Autonomously growing synthetic DNA strands
The Latest on: Self-assembling programmable devices
[google_news title=”” keyword=”self-assembling programmable devices” num_posts=”10″ blurb_length=”0″ show_thumb=”left”]- New class of spongy materials can self-assemble into precisely controllable structureson April 30, 2024 at 8:02 am
A team of researchers led by the University of Massachusetts Amherst has drawn inspiration from a wide variety of natural geometric motifs—including those of 12-sided dice and potato chips—in order to ...
- Quantum forces used to automatically assemble tiny deviceon April 25, 2024 at 1:48 am
The very weak forces of attraction caused by the Casimir effect can now be used to manipulate microscopic gold flakes and turn them into a light-trapping tool ...
- Self-assembling synthetic cells act like living cells with extra abilitieson April 23, 2024 at 5:00 pm
For the new study, scientists from the University of North Carolina at Chapel Hill developed synthetic, self-assembling cytoskeletons ... different peptides or DNA sequences can make for programmable ...
- Self-assembly of complex systems: Hexagonal building blocks are betteron April 3, 2024 at 5:00 pm
Physicists show that the shape of components is a major determinant of how quickly and efficiently complex structures self-assemble. Physicists at LMU Munich show that the shape of components is a ...
- Self-assembly of complex systems: Hexagonal building blocks are betteron April 3, 2024 at 5:00 pm
Gaining a better understanding of the principles and mechanisms of this self-assembly is important for the development of new applications in domains such as nanotechnology and medicine.
- Self-assembly articles from across Nature Portfolioon April 2, 2024 at 4:59 pm
Self-assembly is the process by which an organized ... flexibility enable the fabrication of a high-power-density flexible device. Advances in the development of cytoskeletal-like materials ...
- What are nanobots?on April 3, 2019 at 9:28 am
Researchers are actively exploring the potential of programmable self-assembly properties of nucleic acids to create ... electric, or mechanical device, to such activities as manufacturing. Scale that ...
- Materials Research Science and Engineering Centeron January 17, 2019 at 9:53 am
Self-limiting assembly will be enabled by using DNA origami and polymeric gel particles to create building blocks with programmable shape, anisotropic interactions, and flexibility. These blocks far ...
- What is nanofabrication?on October 22, 2018 at 7:59 am
Efforts to develop purely bottom-up self-assembly methods to create more complex devices typically rely on engineering ... researchers to fold DNA strands into something resembling a programmable ...
- Programmable Logic Devices (PLD) Informationon February 8, 2018 at 12:45 pm
Programmable logic devices (PLD) are designed with configurable logic and flip-flops linked together with programmable interconnect. PLDs provide specific functions, including device-to-device ...
via Google News and Bing News