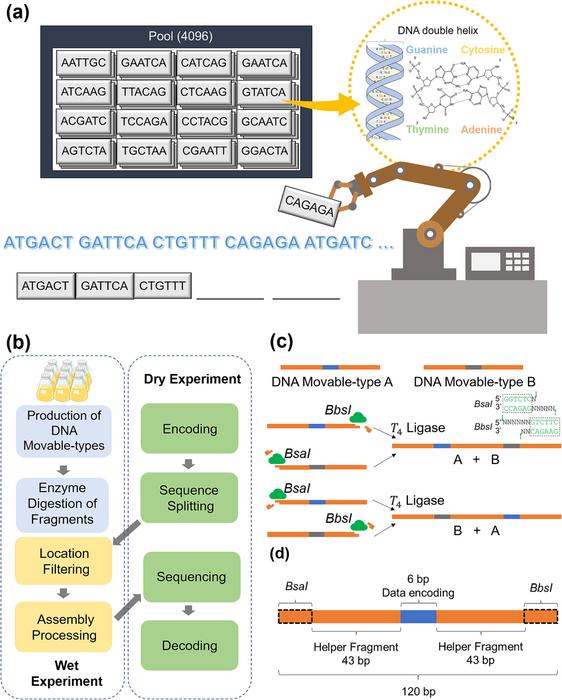
(a) Illustration of the data-writing process of the DNA movable-type storage system. For the data-writing process, high-throughput automation equipment is employed to select the desired DNA movable types and assemble them into corresponding storage units with a length of 408 bp. (b) The overall workflow of the data-writing and -reading processes of the DNA movable-type storage system. (c) Diagram of the ordered assembly of DNA movable types. By selectively digesting either with BbsI or BsaI, the representative DNA movable types A and B can be assembled in a desired order (A-B or B-A) using T4 ligase. Blue and grey areas indicate the data encoding regions. (d) Structure of the DNA movable types. The blue area in the middle stands for the data-encoding region; the two orange modules are helper fragments, which are two randomly generated sequences for improving the ligation efficiency. The two primer binding sets represented by black dotted boxes include two restriction enzyme sites of BbsI and BsaI, respectively. All the DNA movable types have a 6 bp data-encoding region and an overall length of 120 bp. There are 4096 possible sequence combinations for all 6 bp regions, yielding a total of 4096 unique pre-manufactured DNA movable types (a longer data-encoding region can also be applied, in which case the overall number of DNA movable types required to be pre-manufactured will be correspondingly enlarged).
In a groundbreaking study published in Engineering, researchers have developed a revolutionary method for data storage using DNA.
The paper titled “Engineering DNA Materials for Sustainable Data Storage Using a DNA Movable-Type System” introduces a novel approach that utilizes DNA fragments, referred to as “DNA movable types,” for data writing, thereby eliminating the need for costly and environmentally hazardous DNA synthesis.
DNA molecules have long been recognized as green materials with immense potential for high-density and long-term data storage. However, the traditional process of DNA data storage via DNA synthesis has been plagued by high costs and the production of hazards, limiting its practical applications. In response to these challenges, the researchers developed a DNA movable-type storage system inspired by ancient movable-type printing.
The key innovation of this system lies in the use of pre-generated DNA fragments, produced by cell factories, as the basic writing units. These DNA movable types are assembled in a repetitive manner to encode digital information, bypassing the need for de novo DNA synthesis. The process of data writing is achieved by the rapid assembly of these DNA movable types, ensuring a reliable and cost-effective approach to data storage.
The researchers successfully encoded 24 bytes of digital information in DNA using this system and accurately read it back through high-throughput sequencing and decoding. This proof-of-concept demonstration showcases the feasibility and potential of the DNA movable-type storage system.
One of the major advantages of this system is its potential for cost reduction. By utilizing pre-synthesized DNA fragments, the need for expensive DNA synthesis is eliminated. Moreover, the assembly-based data-writing process is entirely biological, making it an environmentally friendly and sustainable technique. The researchers also highlight that this process can be easily parallelized, further increasing its efficiency and scalability.
The DNA movable-type storage system presents a promising route towards economical, sustainable, and environmentally friendly data storage solutions. With the ongoing advancement of high-throughput automation solutions, the researchers expect the data write bandwidth of this system to be substantially improved, meeting the future demands of large data storage.
The potential applications of this innovative system are far-reaching. From archival data storage to cloud computing, this technology has the potential to revolutionize the way we store and access information. By harnessing the power of DNA, researchers are paving the way for a more sustainable and efficient digital data-storage technology.
This groundbreaking research opens up new possibilities for the future of data storage and has significant implications for various industries. With its potential for cost reduction, scalability, and sustainability, the DNA movable-type storage system represents a major step forward in the field of data storage.
Original Article: Revolutionizing data storage: DNA movable-type system paves the way for sustainable data storage technology
More from: Tianjin University
The Latest Updates from Bing News
Go deeper with Bing News on:
DNA movable types
- Millions in federal grants help local police and first responders boost training, buy gear
Tapping federal grants allows IRSC to do a $3 million public safety complex expansion, buys area police and deputies gear, public safety equipment.
- DNA Synthesis Approaches: Will New Methods Stand The Test Of Time?
Both chemical and enzymatic synthesis companies need to continue working to overcome their respective hurdles.
Go deeper with Bing News on:
DNA movable-type storage
- Feed has no items.